OMERO.biobank
Documentation for OMERO.biobank
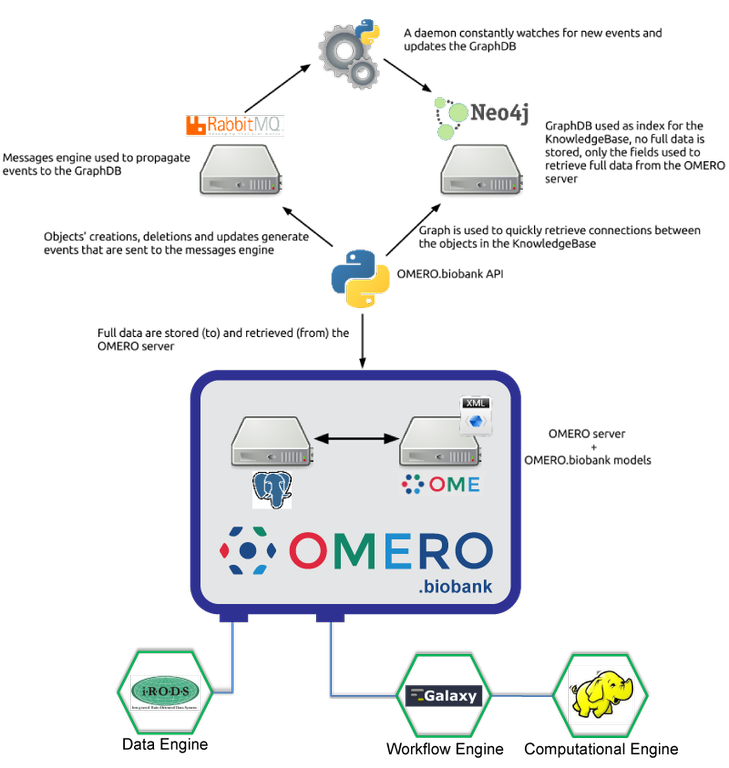
Introduction to OMERO.biobank architecture
OMERO.biobank is part of an ongoing, lab-wide, multi-year project at CRS4 to develop a scalable infrastructure for data intensive biology that natively support reproducible, traceable data processing and analysis with integrated data uncertainty modeling.
The infrastructure uses: a specialized version of OMERO (OMERO.biobank) coupled with a graph database (Neo4j) to model data and the chain of actions that connect them; Galaxy workflows to describe computational processes and Galaxy history mechanism to trap the details that specify a given run instance; Hadoop-based tools to provide scalable computing; and iRODS to decouple dataset logical and physical locations, and to efficiently support inter-institution data sharing.
Biomedical applications
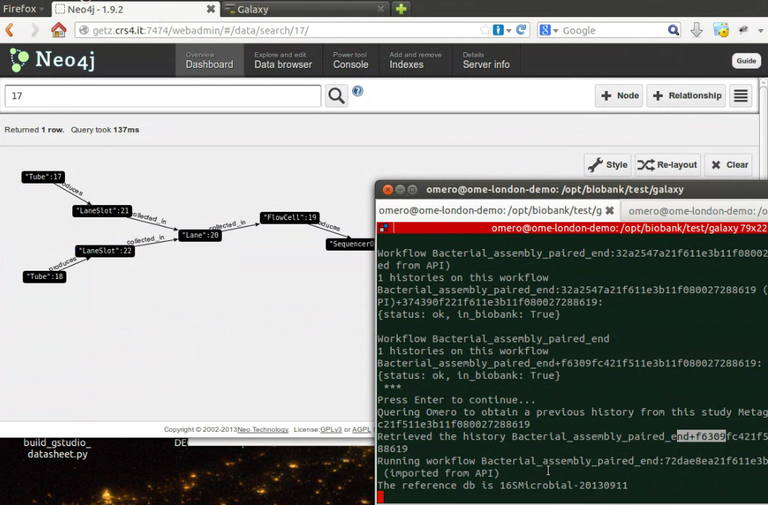
The current version of the infrastructure in production at CRS4 is holding, within others, more than 25,000 microarray datasets (SNPs/CNV and expression), about 3000 Whole genome Re-sequencing, about 700 samples RNA-seq and hundreds of Exome-seq samples. The infrastructure is directly interfaced with CRS4 NGS laboratory and it has been used to build a completely automated NGS computational pipeline, able to process the raw output produced by sequencers into analysis-ready sequence files without human intervention.
Virtual environment
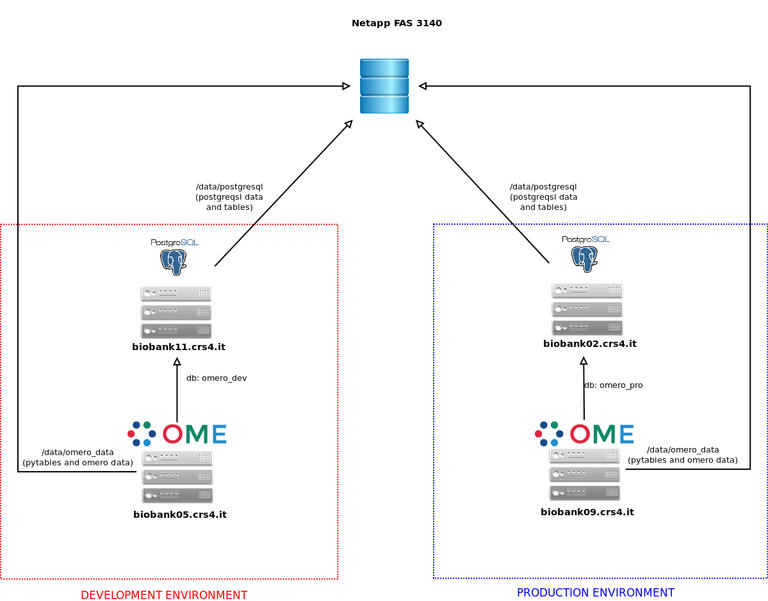
At CRS4, OMERO.biobank is deployed on a virtual environment consisting of two Supermicro 1022G-URF hosts equipped with 16 CPU cores, 32 GB of RAM and 4Gb dual port fiber channel adapters.
We use XenServer free 6.0 as hypervisor and Debian 5/6/7 templates as guests. Storage is provided by a NetApp FAS 3140 file server in High Availability multipath configuration, with a maximum capacity of 54 TB.
OMERO guests use 4 to 8 cores and 8 GB of RAM; the OMERO data dir is mounted on an LVM volume exported by the file server via fiber channel.
PostgreSQL servers run on virtual machines configured in a similar way.
On-going development
The potential applications of OMERO.biobank are not limited to the handling of genetic data. Development has recently been focusing on digital pathology applications. Specifically, working is now on-going to extend OMERO.biobank so that it can be used as a bridge between –omic data produced by the analysis of tissue picked from pathology slides (or paraffin blocks) and the relative morphological information contained in the corresponding digital pathology slides.
Installation instructions
See the OMERO.biobank wiki for installation instructions.
Further information
You may find this presentation given at the 2013 user's meeting a helpful introduction to OMERO.biobank and its future development plans.


