Matlab Analysis
Image Analysis using the OME-MATLAB connector
Introduction
Tom Macura (June 2006)
Updated April 2007
This walk-through illustrates how to convert a legacy image analysis algorithm implemented in MATLAB into an analysis module that can be executed by the OME analysis engine against images and meta-data stored in OME.
Prerequisites
OME must be installed with the optional OME-Matlab connector enabled and you must have a license to execute MATLAB.
Before you do anything else, we strongly recommend that you backup your database. This will allow you to recover should anything unexpected happen. This is especially important if you are performing active development with risky, irreversible operations such as importing analysis modules and chains.
If your database is messed up and you have no backup, your only recourse
will be to reinstall OME after dropping your database. In the process you
will lose all data stored on OME. The command to uninstall OME is
sudo ome admin uninstall
You need to be familiar with the advanced features of the web-ui and ome command-line tool. Please don't proceed without reading those pages first.
Importing and Executing a ready-made Analysis Chain
Once you've got OME up and running, you're ready to try to execute the
Histogram Equalization Test Chain.
This chain takes a gray-scale image and normalizes its contrast by applying the
MATLAB command histeq (from the Image Processing Toolbox). The
HistogramEqualization.ome OME analysis module is a wrapper for
histeq and HistogramEqualizationChain.ome
is a driver feeding pixels into the Histogram Equalization module.
1 Import a test image
If your OME install already contains a dataset of images, you may skip this step and replace the suggested name TestDataset with the name of your actual dataset in subsequent steps.
Import a grayscale image into an OME dataset (e.g. worm.tiff)
[ome.grc.nia.nih.gov:~] tmacur1% ome import -d "TestDataset" /path/to/worm.tiff
2 Import the Analysis Chain and supporting modules
You will need src/xml/OME/Tests/HistogramEqualizationChain.xml and src/xml/OME/Analysis/Filters/HistogramEqualization.ome. These files are provided in the OME distribution but you can also download them using the links above.
Import the module and chain. Because the module is referenced in the analysis chain, the module must be imported first.
[ome.grc.nia.nih.gov:~] tmacur1% ome import src/xml/OME/Analysis/Filters/HistogramEqualization.ome
[ome.grc.nia.nih.gov:~] tmacur1% ome import src/xml/OME/Tests/HistogramEqualizationChain.xml
3 Execute the analysis chain
[ome.grc.nia.nih.gov:~] tmacur1% ome execute -d "TestDataset" -a "Histogram Equalization Preprocessing Chain"
User Inputs:
Histogram Equalization.Histeq NumBins:
This input is optional. New, existing, or skip? [N]/E/S s
Executing Analysis Chain `Histogram Equalization Preprocessing Chain`
1/3: [IN PROGRESS] Executing `Original files`
3/3: [IN PROGRESS] Executing `Histogram Equalization`
3/3: [FINISHED]
Exiting...
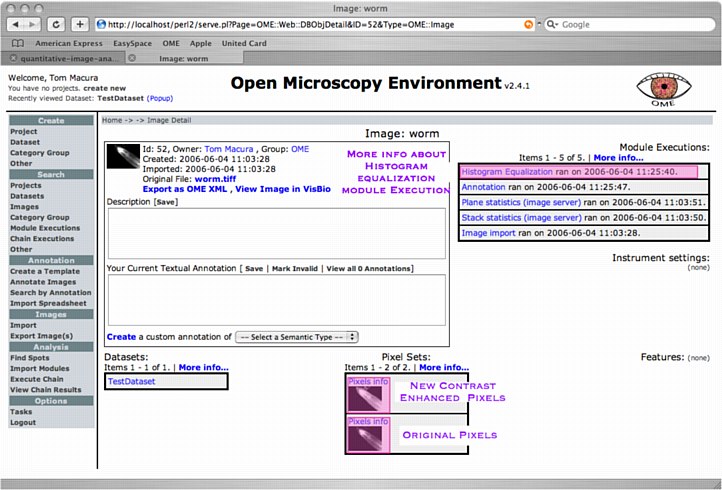
4 Browse Analysis Chain Results
After the Histogram Equalization Chain has been executed, the contrast enhanced pixels are accessible through the Image's Web-UI's ObjectDetail view. You can look into the Histogram Equalization Module's ModuleExecution view to learn additional information e.g. whether there were any warnings from MATLAB and how many seconds it took to run the module.
Writing new Analysis Modules and Analysis Chains
To learn more about how to write a OME analysis module wrapper around a MATLAB function read the Analysis Module XML page, and the MATLAB Handler white-paper.
You will need to copy the module's MATLAB function into the OME .m
file path. This path can be viewed and modified via ome admin configure
[ome.grc.nia.nih.gov:~] tmacur1% sudo ome admin configure
---SNIP---
MATLAB Perl API configuration:
MATLAB User: tmacur1
MATLAB Path: /Applications/MATLAB704
MATLAB Exec: /Applications/MATLAB704/bin/matlab
MATLAB Exec Flags: -nodisplay -nojvm
Config MATLAB for dev?: yes
MATLAB .m files Path: /Users/tmacur1/Desktop/OME-Hacking/OME/src/matlab
---SNIP---
You can write the analysis chain in XML directly.
The steps for executing a ready-made analysis chain and module are exactly the same as executing a module/chain that you've written yourself.
Comments
Please email Tom (tmacura AT nih DOT gov) with any questions/problems you encounter have. Comments are very much appreciated.
Good Luck


