Data Management
Overview of Data Management applications
The OME and OMERO system are made up of several distinct parts, not all of which are necessary for a functional installation.
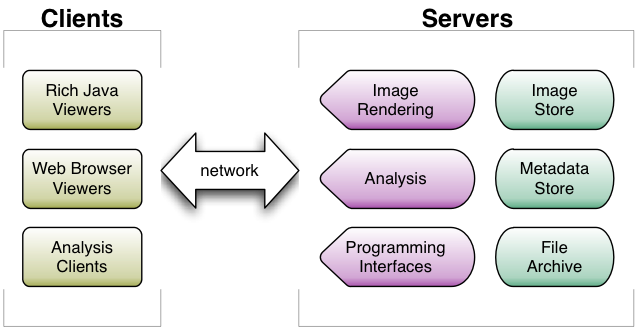
The design follows a standard client-server architecture. A single installation of OME or OMERO will usually be used for an entire laboratory or department. The data and metadata for the images in an OME or OMERO installation are stored on a centralised server. Multiple server processes are used to access the metadata and the binary pixel data. Administrators can choose to place all the server processes on the same machine, or on separate machines, depending on their particular needs and resources.
User access to the data is supported by both traditional user interfaces and programming interfaces. These include:
- light-weight Web-based interfaces, which can be accessed from any computer with a standard Web browser
- rich Java-based client applications providing more functionality than the Web interface, but must be installed separately on each client computer
- a full Java API for access from 3rd party applications like ImageJ and VisBio
- Matlab bindings to facilitate sophisticated image processing and analysis
The data management applications we provide are:
- OME server - The original perl-based OME server project, which implements image-based analysis of cellular dynamics and image-based screening of cellular localization or phenotypes, and includes a fully developed OME-XML Schema language.
- OMERO Platform - The java-based OMERO software project, which currently includes tools for storing, visualizing, managing, and annotating microscopic images and metadata.
- VisBio - The VisBio biological visualization tool. VisBio provides 3D rendered views of multi-dimensional images.


