Automated Image Classification
WND-CHARM Multi-purpose Image Classifier
Tom Macura and Ilya Goldberg (April 2007)
Using pattern analysis to score visual assays
A major limitation of biological visual assays (i.e. microscopy) is the difficulty in obtaining objective quantitative results. Assays often produce pixel data that, either because of the imaging method (e.g. DIC, phase contrast) or sample morphology, is not amenable to traditional computer vision techniques.
Traditional image analysis techniques rely on finding cells, nuclei, or some other "object" of interest, based on an a priori model of the expected object's appearance. This segmentation and model-building step is often very difficult to perform in a robust manner but can be omitted for pattern analysis which can effectively be applied to the entire image.
In a controlled experiment, an assay may need only report whether a given image is like a negative control, or like one or more positive controls. An example of this is high-content-screening (HCS), where one often knows quite specifically what one is looking for, and positive controls are often available that mimic the desired phenotype or morphology. The comparison of a given image to negative and positive controls is an instance of the image classification problem.
Image classification relies on training a classifier using images of controls (classes). The set of images for a given control captures the variability present in the samples and train sthe classifier that this variability is unimportant for classification (i.e. noise). The other classes (each composed of a group of images), are there to train the classifier to find patterns that can differentiate between the classes (i.e. the signal). This process happens automatically. The user only provides several groups of images to define the classes. Once trained, the classifier will assign previously unseen images to one of these defined classes.
The number of images required to train a classifier depends on the variability present within each class and the morphological differences between the classes. A very subtle difference with a lot of variability will require more training images per class than a very obvious difference with little variability. In most cases, even very challenging classification problems can be solved with as few as 50 training images per class. Some with as few as 10. Generally, the more images, the better. Any number of classes can be defined - we've had success with up to 40 classes defined in a classification problem.
What is WND-CHARM?
WND-CHARM is a multi-purpose image classifier developed by Nikita Orlov, Josiah Johnston, Tom Macura, Lior Shamir, and Mark Eckley in Ilya Goldberg's group at the NIA. Our work has shown this classifier to be very robust and accurate in analyzing a variety of image types generally thought inaccessible to automated analysis such as DIC, phase-contrast, complex cytoskeletal and nuclear morphologies, etc. Despite its generality, it is also very accurate - often more accurate than classifiers designed specifically for a particular image classification task. In fact, despite its development for use in cell biology, it is one of the highest scoring algorithms for face recognition, which is significant because this is a very competitive application for image classification. It turns out that once you can do pattern analysis in the wild and wooly world of cell biology, face recognition is a piece of cake.
WND-CHARM consists of two major components: feature extraction (CHARM) and classification (WND-5). CHARM stands for a Compound Hierarchy of Algorithms Representing Morphology. During feature extraction, each image is digested into a set of 1025 image content descriptors (features). The algorithms used to extract these features include polynomial decompositions, high contrast features, pixel statistics, and textures. The features are computed from the raw image, transforms of the image, and transforms of transforms of the image.
After image features have been extracted, there are many classification algorithms that can be utilized: Neural networks, Bayes networks, nearest neighbor algorithms, etc. We developed an algorithm called WND-5 (Weighted Neighbor Distance), which is based on the nearest neighbor method. In our experience, WND-5 is more general and accurate than others we've tried. Additionally, we've found that the class-assignment probabilities it reports can be reliable surrogate measures of image similarity.
WND-5 uses Fisher Discriminants (FD) to score the 1025 features based on their ability to differentiate between classes. The lowest-scoring 1/3 are eliminated. Each of the remaining ~700 features can be thought of as a dimension in a 700-dimensional space where each dimension is weighted by the feature's FD score. Since each image has a value in each of these dimensions, it can be thought of as a point in this space, and the set of training images in a given class would then form a "cloud" of points. The rest of the WND-5 algorithm uses these clouds of points from the training set to assign a previously unseen image to one of the classes. From our work in studying how this classifier performs on different imaging tasks, we attribute its generality and accuracy to the large set of image features, its effective feature selection and weighting algorithm, and its ability to effectively operate in high dimensional space with a limited number of training samples.
Our paper describing WND-CHARM has been submitted for publication in IEEE's Transactions on Pattern Analysis and Machine Intelligence.
WND-CHARM OME Integration
Tom Macura, with Josiah Johnston's help and Ilya Goldberg's guidance, has integrated WND-CHARM into OME. This how-to illustrates how OME and WND-CHARM can be used together to score visual assays.
WND-CHARM is implemented as a set of MATLAB scripts. OME integration of WND-CHARM involved writing XML wrappers called OME Analysis Modules that define the scripts' interfaces in terms of the OME ontology. Lior Shamir has completely re-written WND-CHARM in C/C++ and we are working to integrate the C version of WND-CHARM with OME so MATLAB will no longer be a pre-requisite to doing automated image classification in OME. We are also planning to develop web-ui front-end tools to simplify OME/WND-CHARM integration.
OME integrated WND-CHARM builds on Category-Group/Category classification framework.
This how-to uses the Bangor/Aberystwyth Pollen Image Database as an example that was publicly available from the Informatics Department of Bangor University.
1 Install OME, OME-MATLAB, and WND-CHARM
You need to be familiar with the advanced features of the web-ui and ome command-line tool. Please don't proceed without reading those pages first.
First get the latest version of OME from the CVS repository (OME release 2.6.0 will *not* do). OME must be installed with the optional OME-Matlab connector enabled and you must have a license to execute MATLAB.
Next run the perl script install_classifier.pl. (found in the src/xml/ directory of the OME distribution)
to install the Semantic Types, Analysis Modules, and Chains that constitute the WND-CHARM classifier.
lappytwopy:~/Desktop/OME-Hacking/OME/src/xml tmacur1$ perl install_classifier.pl
\__ OME/Analysis/ROI/DerivedPixels.ome [SUCCESS]. \__ OME/Analysis/ROI/ImageROIConstructors.ome [SUCCESS]. \__ OME/Analysis/Maths/Gradient.ome [SUCCESS]. \__ OME/Analysis/Segmentation/OtsuGlobalThreshold.ome [SUCCESS]. \__ OME/Analysis/Transforms/FourierTransform.ome [SUCCESS]. \__ OME/Analysis/Transforms/WaveletTransform.ome [SUCCESS]. \__ OME/Analysis/Transforms/ChebyshevTransform.ome [SUCCESS]. \__ OME/Analysis/Statistics/EdgeStatistics.ome [SUCCESS]. \__ OME/Analysis/Statistics/ObjectStatistics.ome [SUCCESS]. \__ OME/Analysis/Statistics/CombFirst4Moments.ome [SUCCESS]. \__ OME/Analysis/Statistics/ZernikePolynomials.ome [SUCCESS]. \__ OME/Analysis/Statistics/ChebyshevFourierStatistics.ome [SUCCESS]. \__ OME/Analysis/Statistics/ChebyshevStatistics.ome [SUCCESS]. \__ OME/Analysis/Statistics/HaralickTextures.ome [SUCCESS]. \__ OME/Analysis/Statistics/RadonTransformStatistics.ome [SUCCESS]. \__ OME/Analysis/Statistics/MultiScaleHistograms.ome [SUCCESS]. \__ OME/Analysis/Statistics/TamuraTextures.ome [SUCCESS]. \__ OME/Analysis/Statistics/GaborTextures.ome [SUCCESS]. \__ OME/Analysis/Classifier/FeatureExtractionChain.ome [SUCCESS]. \__ OME/Analysis/Classifier/WND-CHARM-SemanticTypes.ome [SUCCESS]. \__ OME/Analysis/Classifier/WND-CHARM-Prediction.ome [SUCCESS]. \__ OME/Analysis/Classifier/WND-CHARM-Trainer.ome [SUCCESS]. \__ OME/Analysis/Classifier/ImageClassifierPredictionChain.ome [SUCCESS]. \__ OME/Analysis/Classifier/ImageClassifierTrainingChain.ome [SUCCESS].
2 Train WND-CHARM on your image collection
In this step you will import training images into OME, group them into appropriate CategoryGroups/Categories based on image content, and train an image classifier.
Download and extract the Pollen Image Dataset tar archive.
import_test_train_dataset is passed a set of image filenames which it
randomly partitions into two mutually exclusive sets then imports into OME. The --train
subset is 75% and the --test subset is the remaining 25%.
ome dev classifier import_test_train_dataset -d "Pollen_train" --train /path/to/Pollen/*
Use the annotate wizard to infer from the directory structure how pollen images should be grouped by image content.
ome annotate wizard CGC -f pollen_categorization.tsv -s -r /path/to/Pollen/
Take a look at the contents of the pollen_categorization.tsv spreadsheet, then import the spreadsheet to create mass annotations:
ome annotate spreadsheet -f pollen_categorization.tsv
If you look at the Pollen_train dataset in the web-ui you will see it grouped into categories as shown in Figure 1.
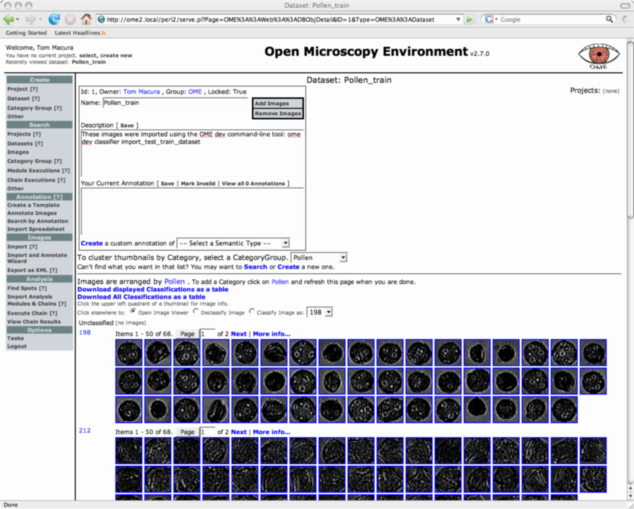
Train a classifier on the training pollen dataset. Expect this command to take about 12 hours to finish running.
ome execute -a "Image Classifier Training Chain" -d "Pollen_train" -s
3 Use WND-CHARM to analyze new images
Import the remaining 25% of the Pollen images to form the test dataset. OME does not know the correct classification for these images. If you look at the Pollen_test dataset using the web-ui (like in Figure 1) you will see the images lacking classifications.
ome dev classifier import_test_train_dataset -d "Pollen_test" --test /path/to/Pollen/*
Use the WND-CHARM classifier (in this example we refer to the classifier trained with ChainExecution ID 3) trained on the Pollen_train dataset to classify images in the Pollen_test dataset. This should run for about 4 hours.
ome dev classifier make_predictions -x 3 -a "Image Classifier Prediction Chain" -d "Pollen_test"
Now look again at the Pollen_test dataset with the web-ui. If all has gone well after selecting the Pollen CategoryGroup under "...Cluster Thumbnails by Category..." you will see the images in this dataset correctly categorized as shown in Figure 2. Once the page has finished loading, you can click the link "Download displayed Classifications as a table" to get a spreadsheet containing all prediction made by WND-CHARM on the test set.
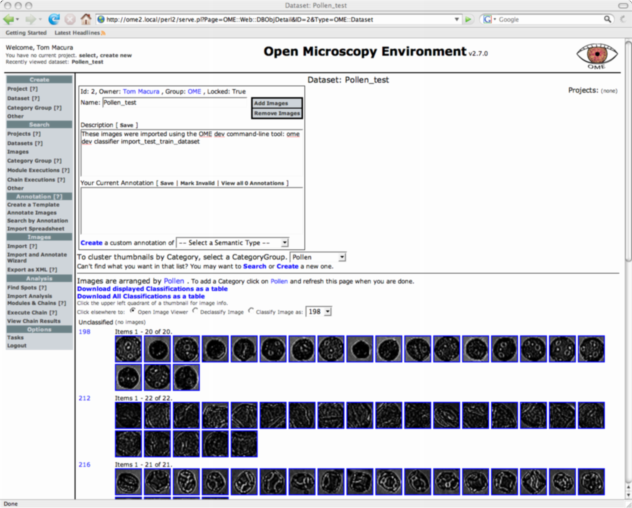
Comments
Please email Tom (tmacura AT nih DOT gov) with any questions/problems you encounter have. Comments are very much appreciated.
N.B.: the behavior of most of the web interface is untested for cases where multiple classifications are stored for a given image (e.g. ground truth and prediction). The dataset view specifically loads the most recent classification of each image, but provides a link to download every classification made on each image. The rest of the web interface will place an image into every category to which it has been classified.


